Model balancing
Model balancing
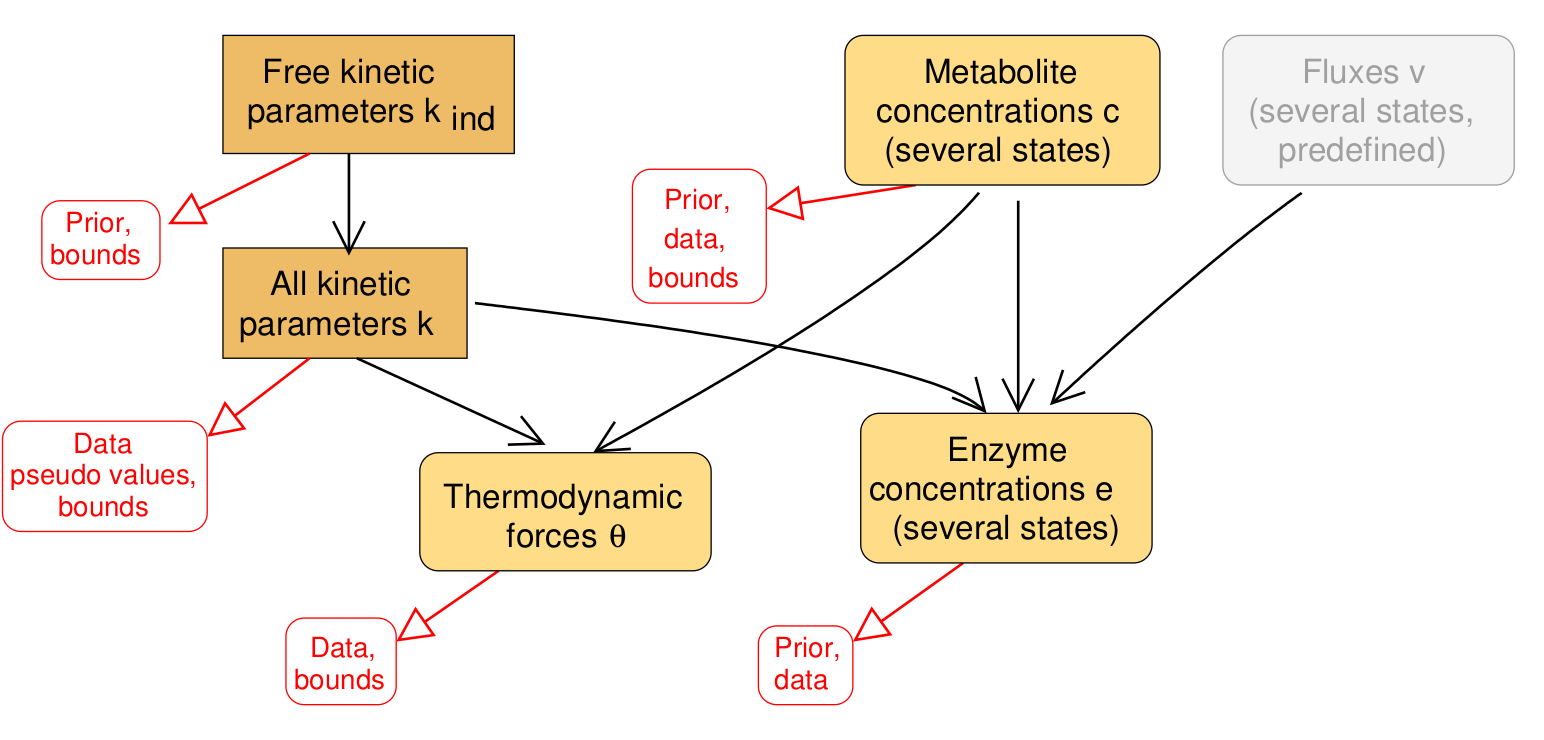
Enzyme kinetic constants in vivo are largely unknown, which limits the construction of large metabolic models. While model fitting, in principle, aims at fitting kinetic constants to measured metabolic fluxes, metabolite concentrations, and enzyme concentrations, the resulting estimation problems are typically non-convex and hard to solve, especially if models are large. Here we assume that metabolic fluxes are known and show how consistent kinetic constants, metabolite concentrations, and enzyme concentrations can be determined simultaneously from data. If one specific term is omitted – a term that penalises small enzyme concentrations – we obtain a convex optimality problem with a unique local optimum. The estimation method with or without this term, called model balancing, applies to models with a wide range of rate laws and accounts for thermodynamic constraints on kinetic constants and metabolite concentrations through thermodynamic forces. It can be used to estimate in-vivo kinetic constants from omics data, to complete and adjust available data, or to construct plausible metabolic states with a predefined flux distribution. As a demonstrative case, we balance a model of E. coli central metabolism with artificial or experimental data. The tests show what information about kinetic constants can be obtained from omics data, and reveal the practical limits of estimating in-vivo kinetic constants.
Reference
If you use model balancing in your work, please cite
my preprint:
Model balancing: in search of consistent metabolic states and in-vivo kinetic constants
Liebermeister W. and Noor E. (2021) bioRxiv doi:10.1101/2019.12.23.887166v2
Code
Matlab and python code is available on github. A brief description of the matlab code can be found here (documentation for all functions is given here). The calculations in the article were run on Matlab version R2019b on Linux.
Contact
Please contact Wolfram Liebermeister with any questions or comments.