Embedding of pathway models into metabolic networks
Main | Models | Code | Matlab help | Article
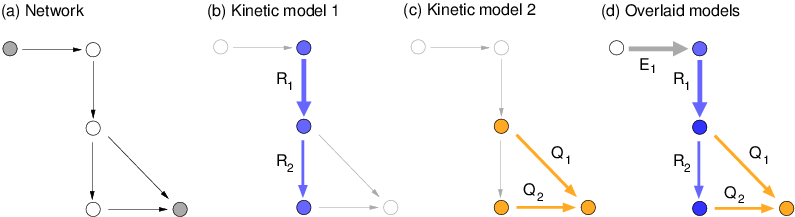
Kinetic pathway models are usually bounded by external metabolites
with fixed concentrations. In reality, pathways are embedded in
larger networks whose dynamics (e.g., a depletion of biosynthetic
precursors) can feed back on the pathways and change their
dynamics. Since network-scale kinetic models are hardly available, I
present an algorithm to embed kinetic pathway models into larger
metabolic network models with simplified kinetics and rate constants
obtained from elasticity sampling.
In the algorithm, the compounds
and reactions are mapped between kinetic and stoichiometric models
and a hybrid model with a metabolic state (characterised by flux
distribution, metabolite levels, and thermodynamic forces) is
constructed. In a second step, the reactions in the stoichiometric
network are equipped with standard rate laws, resulting in a complete
kinetic model with a thermodynamically feasible steady state. The
algorithm allows to control the resulting steady state and guarantees
a thermodynamically feasible model. Two example cases are presented
below.